Over 5,400 different mammalian species exist globally and many have evolved highly distinctive phenotypic traits. The genomic mechanisms underlying these phenotypes are largely unknown but could be relevant for understanding human disease. Accelerated genomic regions may have roles in shaping these important traits. Thus we develop new theories to study accelerated evolution driven by positive selection. Recently, we successfully identified the social enhancer PAS1 that modulates social hierarchy in amniotes.
It can never been underestimated the importance of demography, for example, the demographic histories of human, chimpanzee, cat, fish, insects and plants. However, you cannot infer demography when there are no proper theories. Therefore, by developing new theories, we aim to decipher demographic history of species. This is the beauty of theory as it can reveal evolutionary stories behind each species. These theories are essential for many research fields, for example, precision medicine and conservation.
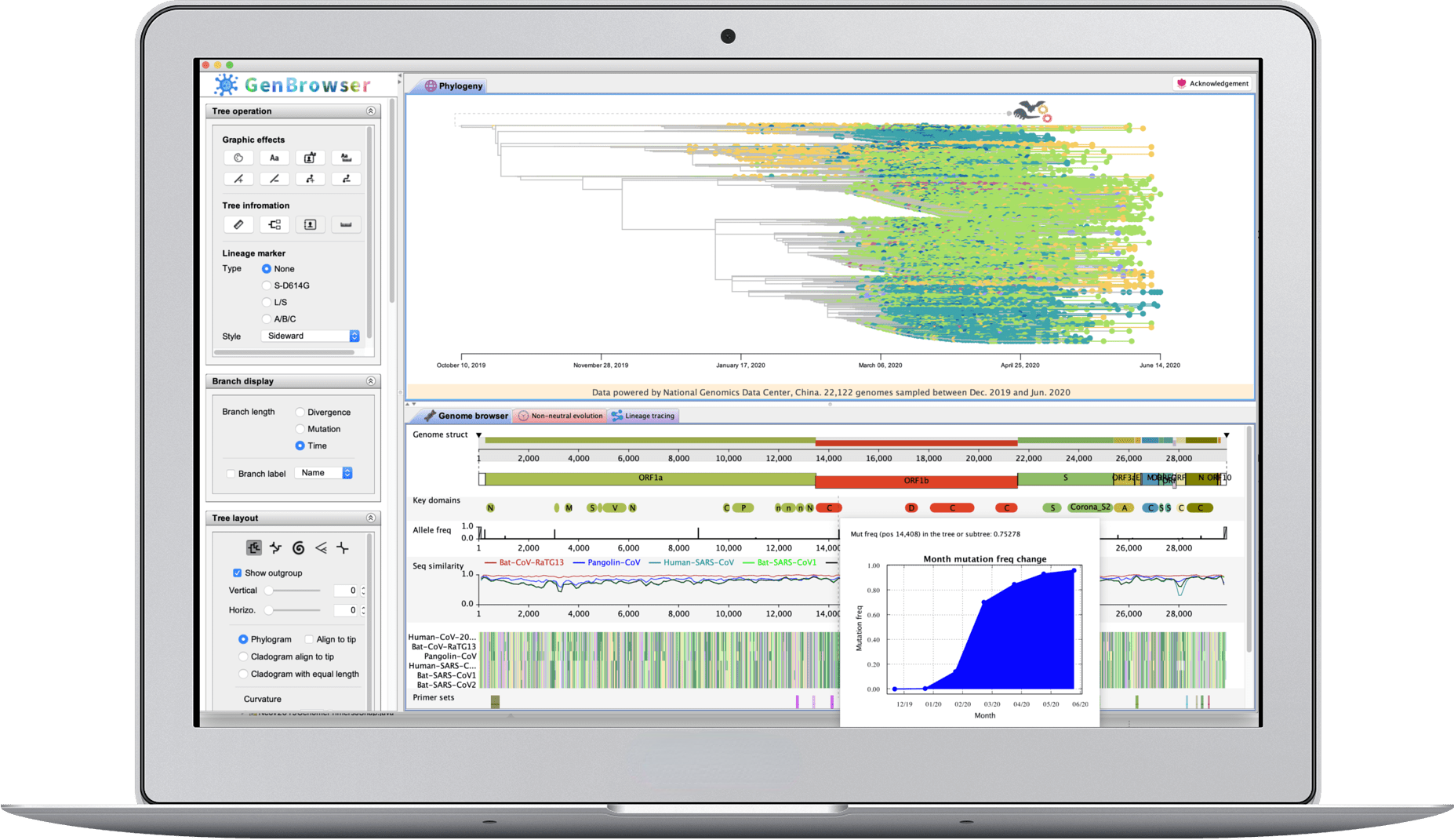
Multiomic and evolutionary analyses have been booming recently, and interactive figures and software are believed to be important for these studies. We combine the advantages of cloud computing and desktop application, and design highly interactive user interfaces and pipelines to bridge the gap between multiomic and evolutionary analyses. Moreover, COVID-19 is spreading across the world. To fight with SARS-CoV-2, the shared enemy of human community, we launch Coronavirus GenBrowser for the public to rapidly access the latest information of SARS-CoV-2 genomes. It can closely monitor the frequency changes of mutations, identify the possible origin of newly emerged strains, and detect the non-neutral evolution of SARS-CoV-2.
Natural selection is one of the important evolutionary forces. By detecting genes and non-coding regions subject to natural selection, it could help us to study the function of genes and non-coding regions, and the interactions between genes and environment.
Our laboratory focuses on developing new theories for fast and efficient methods to detect positive selection. Moreover, since we introduced machine learning (or artificial intelligence) into the field of population genetics ten years ago, we keep exploring possibilities by applying different machine learning methods, for example, deep convolutional neural networks, to detect positive selection. These studies are important for identifying mutations associated with human metabolism and diseases.
Psychobiotics is a term used in preliminary research to refer to live bacteria that, when ingested in appropriate amounts, might confer a mental health benefit by affecting microbiota of the host organism.
Recent advances in nutritional psychiatry point to the importance of the intestinal microbiota to human health and wellbeing. Amazingly, cognitive and emotional processes can be altered by microbes acting through the brain-gut axis. The brain-gut axis provides bidirectional communication between the brain and the gut and includes the metabolically complex intestinal microbiota. However, we need new theories and methods to study why and how gut bacteria can positively or negatively influence the mental process.

LI Haipeng
Room: 329
Tel: 54920460
lihaipeng (at) sinh.ac.cn
2007-present: Professor, Shanghai Institute of Nutrition and Health, China
2006-2007: Postdoctoral Fellow, Institute of Genetics, University of Cologne, Germany
2003-2006: Postdoctoral Fellow, Department of Biology II, University of Munich, Germany
2002-2003: Research Fellow, Human Genetics Center, University of Texas at Houston, USA
2000-2002: Exchange Ph.D. Student, Human Genetics Center, University of Texas at Houston, USA (Co-advisor: Prof. Yun-Xin Fu)
1996-2002: Ph.D. student, Genetics/Zoology, Kunming Institute of Zoology, Chinese Academy of Sciences (Advisor: Prof. Ya-Ping Zhang)
1992-1996: B.S., Zoology, Shandong University, China
Accelerated Evolution | Population Genetics | Bioinformatics | Natural Selection Detection | Psychobiotics

GU Zhili
Group Manager
Room: 335
Tel: 54920467
guzhili (at) sinh.ac.cn

DAI Guangyi
Staff Scientist
Room: 335
Tel: 54920467
daiguangyi (at) sinh.ac.cn

LIN Xin
Ph.D Candidate
Room: 335
Tel: 54920467
linxin2018 (at) sinh.ac.cn

GU Shanshan
Ph.D Candidate
Room: 339
Tel: 54920467
gushanshan2019 (at) sinh.ac.cn

YANG Jianing
Ph.D Candidate
Room: 339
Tel: 54920467
yangjianing2019 (at) sinh.ac.cn

ZHOU Sumei
Ph.D Candidate
Room: 339
Tel: 54920467
zhousumei2020 (at) sinh.ac.cn

ZHANG Jianwei
Ph.D Candidate
Room: 339
Tel: 54920467
zhangjianwei2021 (at) sinh.ac.cn

ZHANG Wanyu
Graduate Student
Room: 339
Tel: 54920467
zhangwanyu2022 (at) sinh.ac.cn

LI Yangyang
Ph.D Candidate
Room: 335
Tel: 54920467
liyangyang2022 (at) sinh.ac.cn

PAN Yichen
Ph.D Candidate
Room: 339
Tel: 54920467
panyichen2023 (at) sinh.ac.cn

ZHOU Yiwen
Ph.D Candidate
Room: 339
Tel: 54920467
zhouyiwen2023 (at) sinh.ac.cn

YAN Fangqi
Staff Scientist
yanfangqi0818 (at) 163.com

XIANGYU Jinggong
Staff Scientist
xiangyujingong (at) picb.ac.cn

MU Hailong
Staff Scientist
mhl_dyx (at) 126.com

LASCOUX Martin
Visiting Scientist
Martin.Lascoux (at) ebc.uu.se

YANG Zongfeng
Ph.D
yangzongfeng (at) picb.ac.cn

LI Junrui
Ph.D
lijunrui (at) picb.ac.cn

LIN Kao
Ph.D
linkao (at) picb.ac.cn

GAO Feng
Ph.D
gaofeng (at) picb.ac.cn

MING Chen
Ph.D
mingchen (at) picb.ac.cn

WANG Yuting
Ph.D
wangyuting (at) picb.ac.cn

YANG Jinjin
Intern
yangjinjin2009 (at) gmail.com

HUANG Hongwei
Intern
huanghw1111 (at) 163.com

HU Wangjie
Ph.D
wangjie.hu (at) mssm.edu

HAO Ziqian
Ph.D
haoziqian (at) sdfmu.edu.cn

DU Pengyuan
Ph.D
dupengyuan (at) sdfmu.edu.cn

LIU Guopeng
Ph.D
guopeng.liu (at) abogenbio.com

YU Dalang
Ph.D
yudalang (at) ucas.edu.cn

SHI Yi
Ph.D
shiyi2017 (at) sinh.ac.cn



 [Green open version, supplementary materials, and introduction slides]
[Green open version, supplementary materials, and introduction slides]